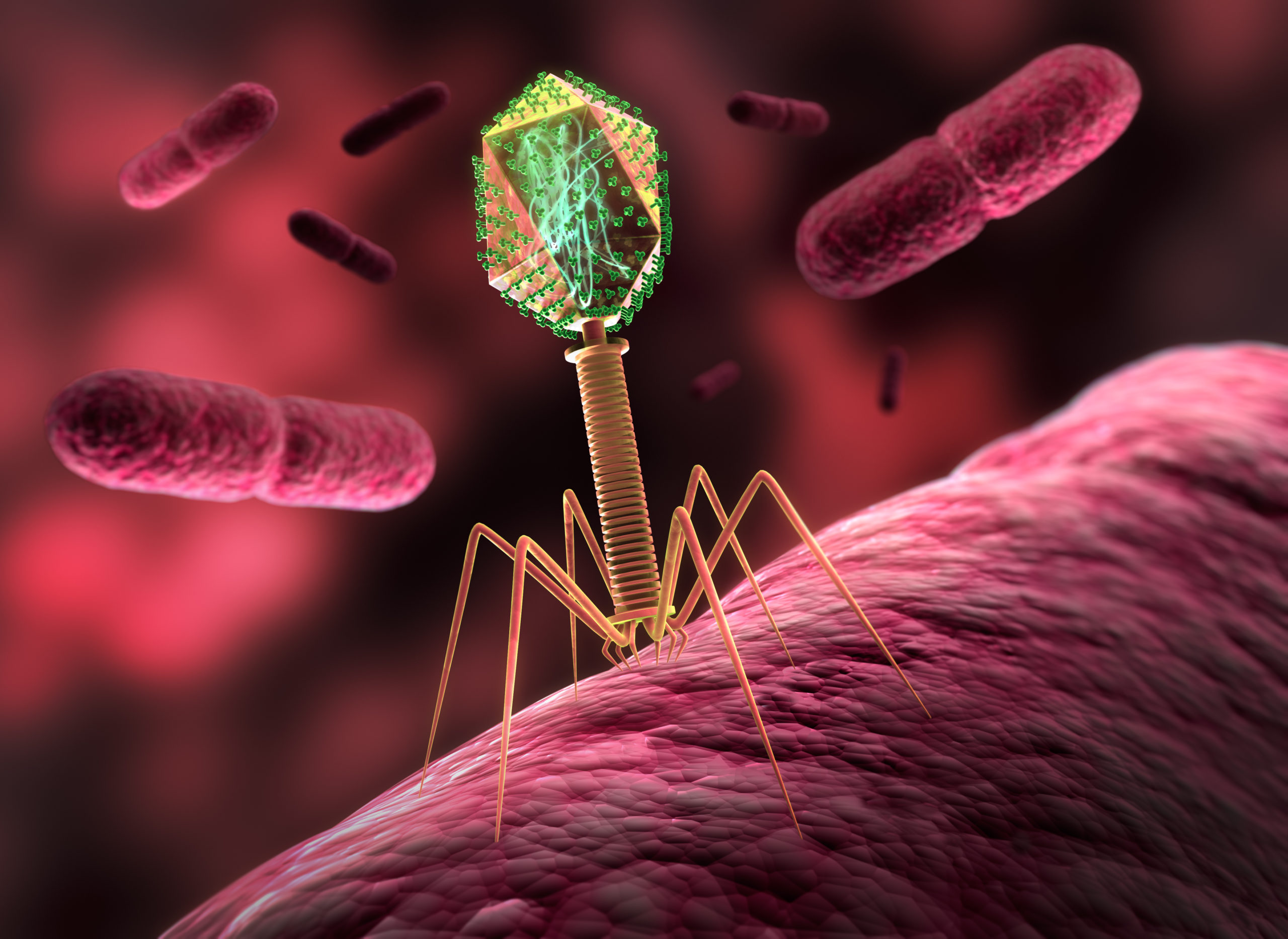
Just as viruses need our cells in order to replicate, bacteriophages need the bacteria they infect in order to replicate. Different bacteriophages target different bacteria. Typically, bacteriophages attach to receptors on the surface of certain bacteria and infect them by injecting their genetic material. “Virulent” phages have a lytic cycle, during which they use the energetic resources of the infected bacteria to replicate their genetic material and form new bacteriophages (up to 1,000 new phages per bacterium), which are then released by destroying the bacteria. So-called “temperate” phages have a lytic cycle but also a lysogenic cycle, during which they replicate along with the infected bacteria without killing them.
The ability of bacteriophages to selectively destroy certain bacteria has long been known. During the 20th century, researchers used bacteriophages as drugs to treat bacterial infections such as dysentery. However, at the time, this approach was considered less reliable than antibiotics. As a result, most countries, with the exception of those in the Soviet bloc, abandoned bacteriophage therapy in favor of antibiotics. Today, while bacteriophage therapy is not authorized in Europe or the US, bacteriophages are used by the food industry to make sure that products intended for consumption do not contain certain bacteria At the same time, the problem of bacterial resistance to antibiotics has revived interest in a therapeutic approach involving the use of bacteriophages2.
Concerning the gut microbiome, bacteria have been studied more than the viruses that infect them because it is much easier. Indeed, bacteria is characterized largely based on the fact that some bacterial genes are very similar between bacteria of the same species but distinct between bacteria of different species 3-4. However, compared to bacteria, bacteriophage genes have a much higher variability 5. In other words, two “identical” bacteriophages are often much more different genetically than two “identical” bacteria. For example, in 97% of cases there is no homology between the genomic sequences of all identified bacteriophages (several thousand) 6. And while it is possible to name the bacteria present in a given sample based on all the DNA present in it, this is not possible with bacteriophages. Consequently, 75–99% of bacteriophage DNA sequences do not match any known viral DNA sequence 7.
Despite these difficulties, methods have been developed to characterize the bacteriophages in the gut, and the first human virome (or “phageome”) was published in 2003 8. It is estimated that there are about 108 to 109 bacteriophages per gram of feces, i.e. about 1012 bacteriophages in the human gut 5,9. Like the bacteria in the gut, the gut bacteriophage composition fluctuates during the first two years of life and then remains relatively stable over time, even as it is influenced by diet 10-14. The majority of this gut virome is identical between healthy individuals, composed mainly of virulent bacteriophages such as Caudovirales and Microviridae, and only a minority of bacteriophages is unique to each individual 15,16.
In the end, why should we care if bacteria get infected by bacteriophages in our gut? Well, quite simply because bacteriophages act as regulators of the bacterial populations they infect. As they pass from bacteria to bacteria, bacteriophages introduce new genes, contributing to the diversification of bacterial populations 17. And bacteriophages can have harmful or beneficial effects on our health. An example of a detrimental effect is that bacteriophages can carry toxin-coding genes from pathogenic bacteria to non-pathogenic bacteria. An example of a beneficial effect is that colonization of the mucus layer overlying the intestinal epithelium by bacteriophages provides additional protection, on top of immune defenses (antibodies and antibacterial peptides), against invasive bacteria 18-19.
If viruses contribute to the bacterial balance that is so essential to maintaining good health, are viral imbalances detected in diseases related to the gut microbiome? The answer is yes. The bacteriophage composition in patients with chronic inflammatory bowel disease (including Crohn’s disease and ulcerative colitis) is different 15,20. While these patients have a reduced bacterial diversity, they have greater bacteriophage (in particular Caudovirales) diversity and an increase in the ratio of temperate to virulent bacteriophages compared to healthy individuals 21,22.
The likely role of bacteriophages in regulating bacterial populations in the gut and the fact that bacteriophage composition is associated with gut pathology points to a possible broader role of bacteriophages. The astounding efficacy of fecal microbial transplant (FMT) in treating Clostridium difficile infections in the gut has raised hopes that such an approach could potentially be effective for other intestinal pathologies 23–26. FMT is generally thought to work by replacing a defective bacterial microbiome with a healthy one 27. However, the transfer of fecal matter generally does not involve transferring only gut bacteria but everything else as well. Indeed, one study showed that FMT works just as well if the bacteria are removed from the transferred feces to the recipient, raising the question of the role of gut bacteriophages in the effectiveness of this approach 28 Bacteriophages may also play a role in the response to cancer treatments. Cyclophosphamide, a molecule widely used in chemotherapy, induces the passage of the gut bacterium E. hirae from the gut to the lymphoid organs, where it provokes an immune response that can target tumor cells. Laurence Zitvogel’s group demonstrated that it was not these bacteria that activated a specific anti-cancer immune response but a bacteriophage located inside these bacteria, whose molecules were very similar to those expressed by cancer cells 29.
The gut microbiome is therefore much more than just the sum of the bacteria in the gut, and the development of new tools and methods will likely lead to a better understanding in the coming years of the impact that gut bacteriophages may have on health.
References
- W. B. Whitman, D. C. Coleman et W. J. Wiebe. “Prokaryotes : the unseen majority”. In : Proc Natl Acad Sci U S A 95.12 (juin 1998), p. 6578-6583.
- D. M. Lin, B. Koskella et H. C. Lin. “Phage therapy : An alternative to antibiotics in the age of multi-drug resistance”. In : World J Gastrointest Pharmacol Ther 8.3 (août 2017), p. 162-173.
- M. H. Fraher, P. W. O’Toole et E. M. Quigley. “Techniques used to characterize the gut microbiota : a guide for the clinician”. In : Nat Rev Gastroenterol Hepatol 9.6 (mar. 2012), p. 312-322.
- D. J. Lane et al. “Rapid determination of 16S ribosomal RNA sequences for phylogenetic analyses”. In : Proc Natl Acad Sci U S A 82.20 (oct. 1985), p. 6955-6959.
- M. B. Dion, F. Oechslin et S. Moineau. “Phage diversity, genomics and phylogeny”. In : Nat Rev Microbiol 18.3 (mar. 2020), p. 125-138.
- T. N. Mavrich et G. F. Hatfull. “Bacteriophage evolution differs by host, lifestyle and genome”. In : Nat Microbiol 2 (juil. 2017), p. 17112.
- V. Aggarwala, G. Liang et F. D. Bushman. “Viral communities of the human gut : meta- genomic analysis of composition and dynamics”. In : Mob DNA 8 (2017), p. 12.
- M. Breitbart et al. “Metagenomic analyses of an uncultured viral community from human feces”. In : J Bacteriol 185.20 (oct. 2003), p. 6220-6223.
- A. N. Shkoporov et C. Hill. “Bacteriophages of the Human Gut : The “Known Unknown” of the Microbiome”. In : Cell Host Microbe 25.2 (fév. 2019), p. 195-209.
- E. S. Lim et al. “Early life dynamics of the human gut virome and bacterial microbiome in infants”. In : Nat Med 21.10 (oct. 2015), p. 1228-1234.
- A. Reyes et al. “Viruses in the faecal microbiota of monozygotic twins and their mothers”. In : Nature 466.7304 (juil. 2010), p. 334-338.
- S. Minot et al. “Rapid evolution of the human gut virome”. In : Proc Natl Acad Sci U S A 110.30 (juil. 2013), p. 12450-12455.
- A. N. Shkoporov et al. “The Human Gut Virome Is Highly Diverse, Stable, and Individual Specific”. In : Cell Host Microbe 26.4 (oct. 2019), p. 527-541.
- S. Minot et al. “The human gut virome : inter-individual variation and dynamic response to diet”. In : Genome Res 21.10 (oct. 2011), p. 1616-1625.
- P. Manrique et al. “Healthy human gut phageome”. In : Proc Natl Acad Sci U S A 113.37 (sept. 2016), p. 10400-10405.
- A. N. Shkoporov et al. “Reproducible protocols for metagenomic analysis of human faecal phageomes”. In : Microbiome 6.1 (avr. 2018), p. 68.
- R. Sausset et al. “New insights into intestinal phages”. In : Mucosal Immunol 13.2 (mar. 2020), p. 205-215.
- G. M. F. Almeida et al. “Bacteriophage Adherence to Mucus Mediates Preventive Protection against Pathogenic Bacteria”. In : mBio 10.6 (nov. 2019).
- J. J. Barr et al. “Bacteriophage adhering to mucus provide a non-host-derived immunity”. In : Proc Natl Acad Sci U S A 110.26 (juin 2013), p. 10771-10776.
- T. Zuo et al. “Gut mucosal virome alterations in ulcerative colitis”. In : Gut 68.7 (juil. 2019), p. 1169-1179.
- J. M. Norman et al. “Disease-specific alterations in the enteric virome in inflammatory bowel disease”. In : Cell 160.3 (jan. 2015), p. 447-460.
- A. G. Clooney et al. “Whole-Virome Analysis Sheds Light on Viral Dark Matter in Inflam- matory Bowel Disease”. In : Cell Host Microbe 26.6 (déc. 2019), p. 764-778.
- J. S. Bakken. “Fecal bacteriotherapy for recurrent Clostridium difficile infection”. In : Anae- robe 15.6 (déc. 2009), p. 285-289.
- E. van Nood et al. “Duodenal infusion of donor feces for recurrent Clostridium difficile”. In : N Engl J Med 368.5 (jan. 2013), p. 407-415.
- T. J. Borody et A. Khoruts. “Fecal microbiota transplantation and emerging applications”. In : Nat Rev Gastroenterol Hepatol 9.2 (déc. 2011), p. 88-96.
- L. P. Smits et al. “Therapeutic potential of fecal microbiota transplantation”. In : Gastroen- terology 145.5 (nov. 2013), p. 946-953.
- A. Khoruts et M. J. Sadowsky. “Understanding the mechanisms of faecal microbiota trans- plantation”. In : Nat Rev Gastroenterol Hepatol 13.9 (sept. 2016), p. 508-516.
- T. Zuo et al. “Bacteriophage transfer during faecal microbiota transplantation in Clostridium difficile infection is associated with treatment outcome”. In : Gut 67.4 (avr. 2018), p. 634-643.
- A. Fluckiger et al. “Cross-reactivity between tumor MHC class I-restricted antigens and an enterococcal bacteriophage”. In : Science 369.6506 (août 2020), p. 936-942.
Author

Julien Verdier